Genes and heart disease: Finally making the link
Polygenic risk scores — a patient’s chance, based on tiny DNA variants, of developing cardiovascular disease, breast cancer and more — are coming to clinics. But there are kinks to iron out and accuracy remains an issue.
Support sound science and smart stories
Help us make scientific knowledge accessible to all
Donate today
What are your chances of having a heart attack in the next decade? Knowing that would likely inform your health and lifestyle choices. Your physician can feed factors including your age, cholesterol levels and blood pressure into an equation that spits out an estimated risk level for the next 10 years: High? Low? Somewhere in between? If the risk is high, your doctor can recommend changes: Eat a healthier diet, exercise more, etc.
But these predictions are far from perfect. Studies suggest that most of these calculators tend to overestimate risk, by up to 154 percent, and can also underestimate. That’s partly because the calculators are missing a lot of relevant inputs. Crucially, in this modern genetic age, they leave out known but poorly understood genetic factors that are responsible for about half of a person’s risk of cardiovascular disease.
It may sound surprising that we’re not further along in our use of genes to predict health. After all, it’s been more than 20 years since the 2003 release of the first human genome sequence. That data trove was expected to usher in a new era in which key genes would predict risk and allow for personalized medical recommendations. Even before the human genome sequence, for example, we knew that having a gene variant associated with Huntington’s disease meant the neurological condition would eventually arise. And we knew that mutations in the two BRCA genes boosted breast cancer risk so significantly that some women opt to protect themselves with preemptive mastectomies.
But here’s the rub: It turns out that big-impact genes like those are few and far between. And to the surprise of the genome sequencers, only 2 percent of the genome actually codes for proteins, while many variants linked to disease are in the other 98 percent, once called “junk DNA.” Scientists now know much of that DNA is far from junk, but instead helps to control how the protein-coding genes are used by cells. And they know that much of the risk for conditions like cancer and heart disease results from tiny variations in many far-flung genetic letters that each individually have minuscule impacts. Some of the variations might protect against disease a little bit; others might make it a smidge more likely. If doctors could add up all those little bitty factors, all those tiny pluses and minuses, they might be able to offer much better estimates for disease risk. Still, it’s been devilishly difficult to figure out all the associations.
Today, the net value of all those tiny genetic influencers, called a polygenic risk score, is nearing clinical use. Progress is farthest along for cardiovascular disease, and breast cancer predictors are also incorporating small-effect variants, while the usefulness of the risk scores for other cancers is being assessed. Polygenic risk scores for conditions like asthma, obesity and diabetes are also under investigation across a network of research sites. Outside the standard health care sector, companies such as 23andMe are already offering paying customers a number of risk scores and associated health advice.
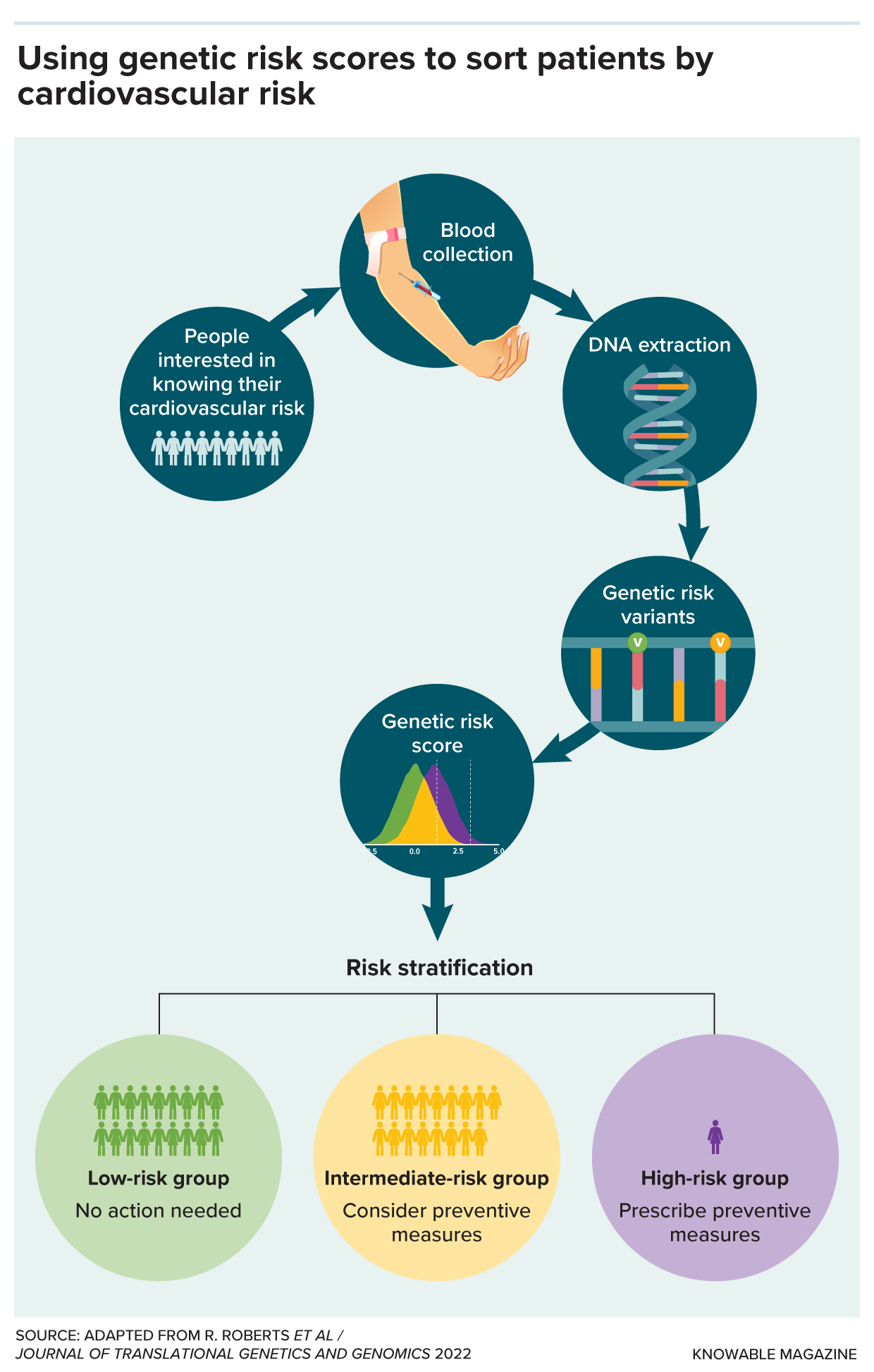
Here’s how polygenic risk scores for cardiovascular disease can be used in a medical setting. People interested in understanding how their genes affect their risk for cardiovascular disease have a blood sample taken. Scientists extract the DNA from their blood and identify variants in the genome that are associated with higher or lower risk. From all those variants, they can calculate a polygenic risk score. People at low risk probably require no further attention. People at high risk will likely be prescribed interventions, such as statins. People in the intermediate group might consider protective measures, such as dietary changes.
Significant hurdles remain. For many diseases, scientists need more data to identify all the relevant small-effect genes. And even in cases like heart disease where that gene list is relatively comprehensive, risk scores can be confusing — for both patients and care providers. To integrate the scores broadly into medical practice, researchers must work out how to effectively and accurately communicate risk and how to apply the information to improve a person’s health outlook.
Another major problem is that most genome studies have been done on people of European ancestry, which results in less accurate risk scores for people of other ethnicities. Scientists are now focused on collecting more genetic data to improve polygenic risk scores for people of varied descent.
“It’s not appropriate to roll them out only for one ethnicity, or to apply [them] to all ethnicities knowing that it doesn’t work well, so this is really an issue of justice,” says Russ Altman, a computational geneticist at Stanford University in California.
Thus, for most health conditions, creating clinical-grade, reproducible scores that physicians will trust and adopt will take more work, researchers say — and that work is underway.
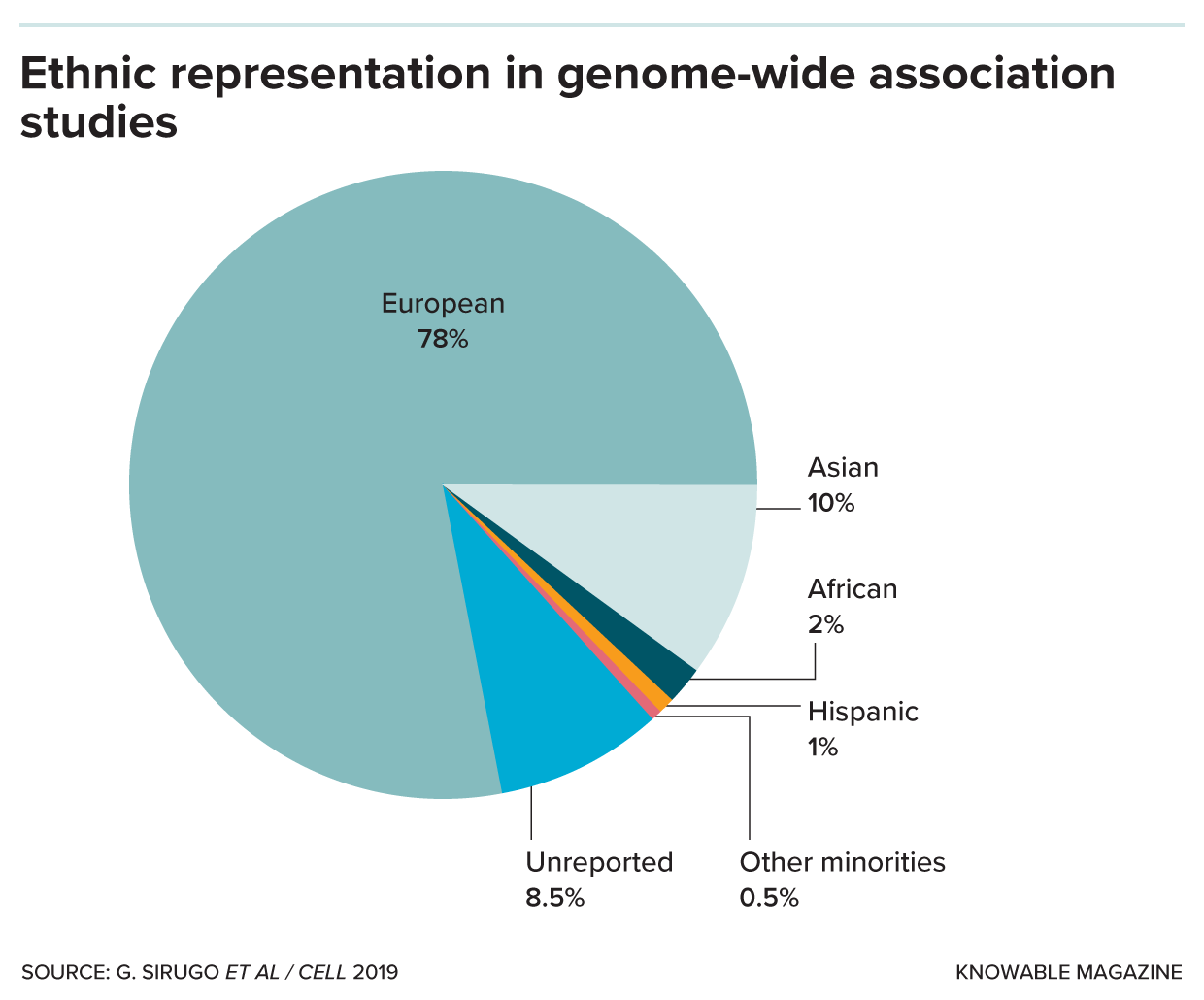
Genome-wide association studies (known as GWAS) are used to create the algorithms for polygenic risk scores. But they are based on a subject pool that is heavily European, despite the fact that Europeans make up only about 16 percent of the world’s population.
Beyond the pea prototype
Polygenic risk scores are based on genetics that is different from the kind many of us learned in high school. For the monk Gregor Mendel and his pea plants, a single gene controlled the pea’s color: One variant of the gene made it green, another made it yellow, and so on for other pea traits that he studied. Some human diseases work in the same way: Sickle cell disease, for example, or cystic fibrosis or hemophilia result from single-gene mutations.
But many common traits and conditions have a much more complex inheritance pattern. For things like height, cardiovascular health and many cancers, the genetic lottery depends not on one or a few all-powerful genes, but on thousands, even hundreds of thousands, of tiny DNA variations among people. A variation might be a single change in a genetic letter, an A substituted by a T, for example. Or it might be a small insertion or deletion in that string of DNA letters. One change might raise disease risk by, say, 0.001 percent. Another might lower it by 0.005 percent. And so on. Polygenic risk scores are algorithms that sum up all those tiny factors.
Scientists find correlations between genetic variations and diseases via what are called genome-wide association studies, which examine the genomes of hundreds or thousands of people, along with their medical histories, and then ask whether a given genetic variation tends to coincide with a health condition. Now, after two decades of genome-wide association studies, scientists know a lot — and the calculation of polygenic scores can finally commence.
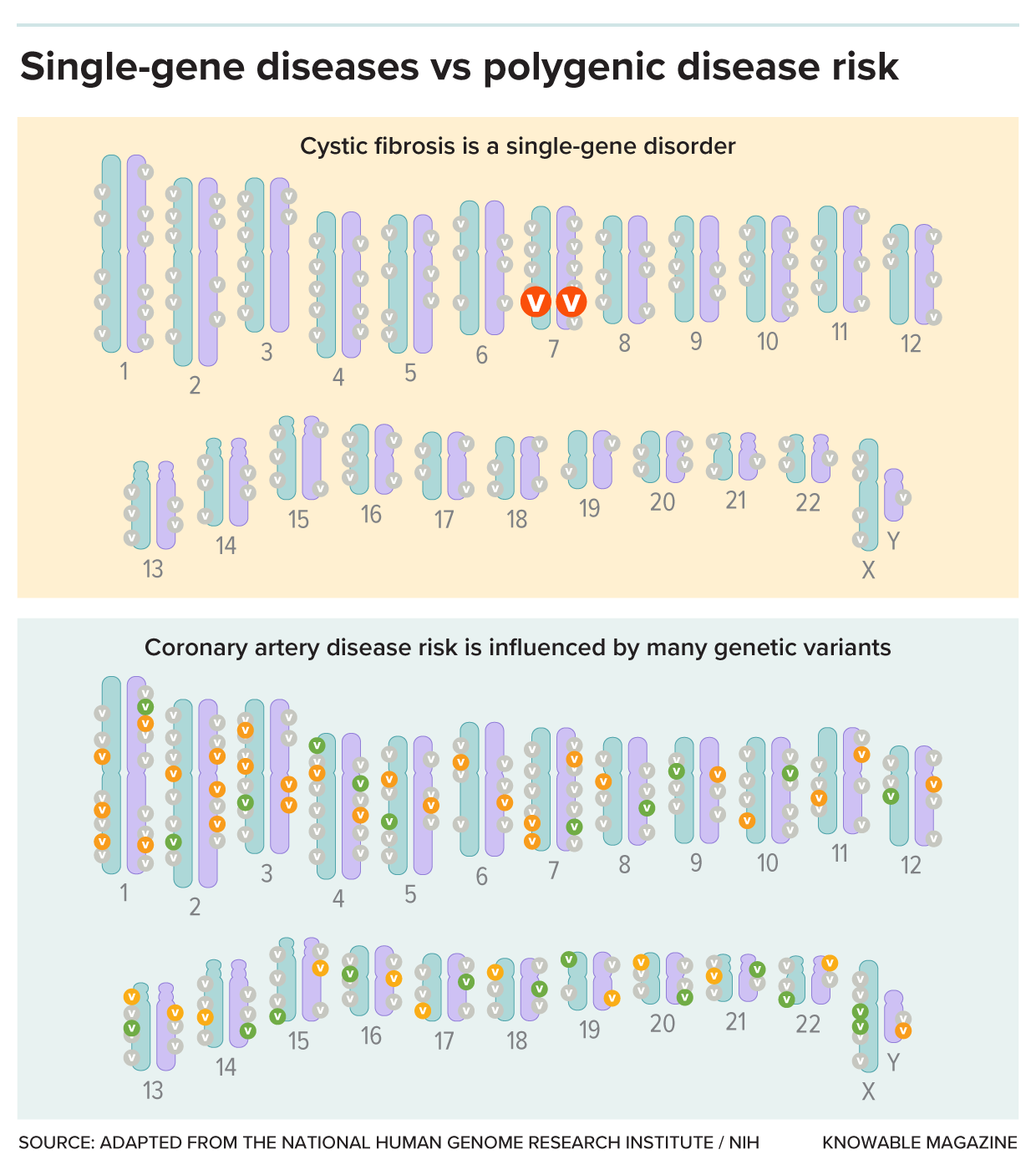
Nobody’s genome is identical, and some genetic variants cause a disease or raise the risk for one. In the case of single-gene disorders (top), such as cystic fibrosis, a variant in a single gene is enough to cause disease. For polygenic conditions (bottom), it’s more complicated. Many genetic variants are involved, each with small effects. Some variants (green) will be a little bit protective, while others (orange) will add a bit to overall risk. Polygenic risk scores are the net values of all those variants.
There’s a caveat to genome-wide association studies: Just because a variant tends to correlate with a disease does not necessarily mean the variant causes the disease. “They’re just tags,” says Alicia Martin, a human geneticist at the Broad Institute of MIT and Harvard in Cambridge, Massachusetts, and Massachusetts General Hospital in Boston. The tag might merely be located close to the actual risk-raising culprit on a chromosome, such that they tend to be inherited together. But for polygenic risk scores, this doesn’t matter. All researchers need to know is that the presence of that tag means risk for the disease goes up or down.
Researchers stress that polygenic risk scores are not genetic destiny the way that, say, the Huntington’s or sickle-cell gene is. Risk is generally modifiable. If a person’s genetic risk for heart disease is high, they could exercise and eat right and very likely lower their overall risk. Conversely, a low genetic risk shouldn’t be taken as a license to eat bacon at every meal: Poor health choices could still put the heart at risk.
The promise of polygenic risk scores is to identify people at high risk so that they can take action to protect themselves or get regular screening for diseases, says Bertram Koelsch, a biomedical engineer and director of product research and development at 23andMe in Sunnyvale, California. The company’s reports include nearly 40 different risk scores for conditions including melanoma, gestational diabetes and migraines.
Some companies even claim to use polygenic risk scores to screen embryos for couples planning pregnancies, though many experts say it’s inappropriate to do so.
Still, these commercial scores are a “black box” because their algorithms are proprietary and it’s often unclear which populations they were tested and validated in, says Aniruddh Patel, a cardiologist at Massachusetts General Hospital. (Koelsch says 23andMe does train and test its scores in people from diverse backgrounds.) Telling someone if they’re in the highest one-fifth or lowest one-fifth of individuals for heart health risk could be useful, and the advice — for instance, to stop smoking — isn’t going to hurt anyone, says Alexander Hatoum, a behavior geneticist at Washington University in St. Louis. But, he adds, “most disorders aren’t ready.”
For polygenic risk scores to reach widespread use, physicians will want dedicated training and official guidelines that tell them how to counsel patients about their scores, Patel says. A cardiologist like him, for example, would want clear guidance from the American Heart Association or the American College of Cardiology.
And making risk score reports understandable to clinicians and patients is not straightforward. In a 2022 study, for example, Massachusetts General Hospital researchers tested mock polygenic risk score reports with 25 subjects acting as “patients” and 21 primary-care providers. Many patients didn’t understand what the numbers meant. For example, when told they were in the 95th percentile of risk, meaning their risk was higher than 95 percent of people, many interpreted it to mean their absolute risk of getting the disease was 95 percent. Even two primary care providers made this mistake.
It’s also important to consider the average risk for a condition, Martin says. Take something like thyroid cancer: There are about 43,720 new cases in the United States each year, affecting 1.2 percent of people over their lifetime. Even if DNA variants double or triple a person’s risk, it’s still a small risk.
That’s a lot of nuance to take in at a doctor’s appointment.
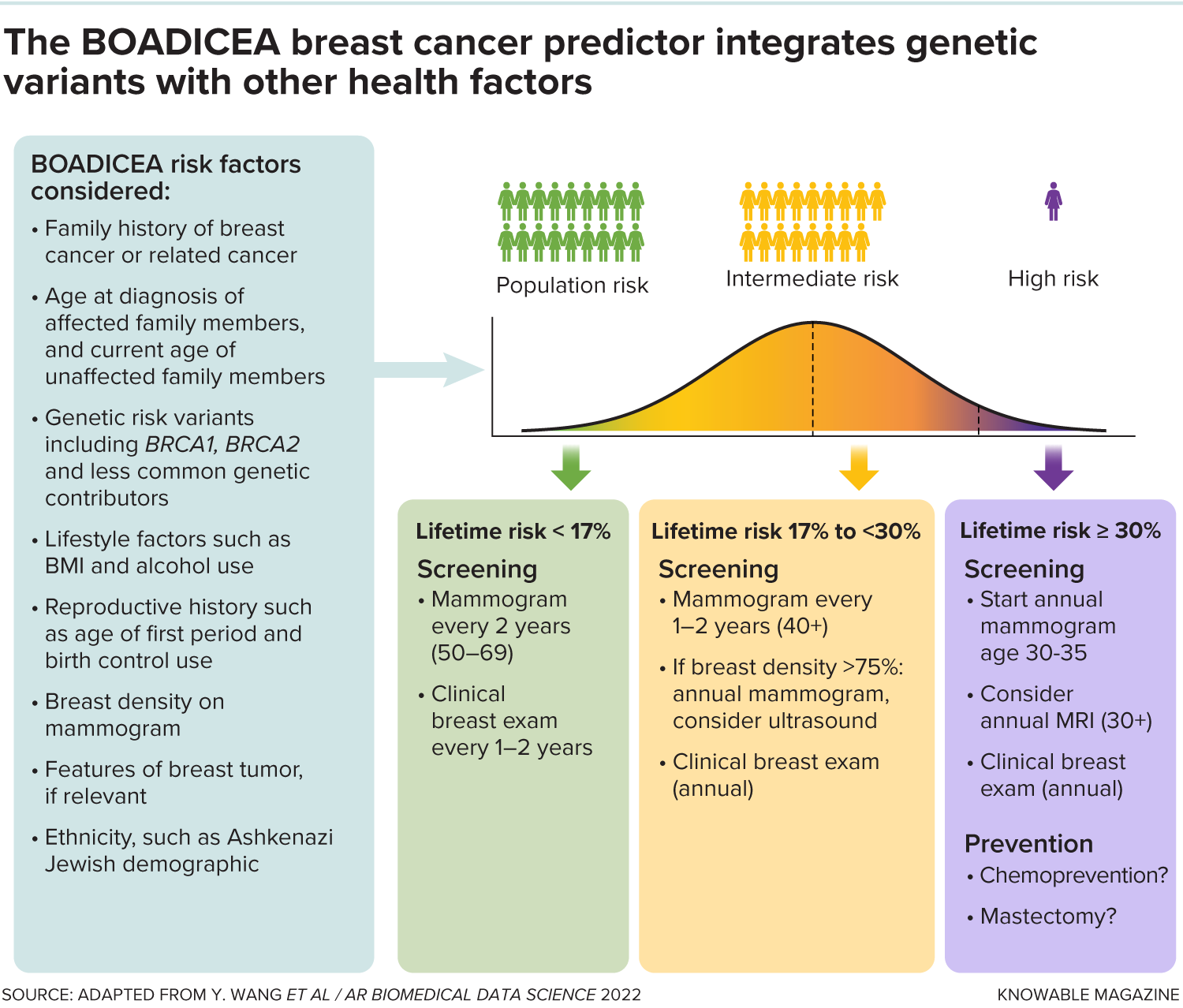
The BOADICEA breast cancer prediction algorithm incorporates a variety of factors, including genetic variants, to calculate a person’s lifetime risk for breast cancer. Most people will end up with a lifetime risk that is low (the bottom 17 percent of the population) or intermediate (between 17 percent and 30 percent of the population). The higher the calculated risk, the more screening and preventive measures are recommended.
Beyond proper counseling, there’s also the cost. Identifying the variants in an individual’s genome can cost as little as $40, says Patel, but the expert interpretation or physician training will take money, as will setting up secure servers to store extensive genetic data — about 100 gigabytes per genome. “In order for it to come to a clinic near you, insurance companies have to buy in,” says Patel, who wrote about polygenic risk scores for coronary artery disease in the 2023 Annual Review of Medicine. Scientists therefore need to prove that the risk scores provide good value for the investment.
They’re getting the closest to that with cardiovascular disease.
In a 2021 study, for example, researchers in the United Kingdom used data from the UK Biobank, a huge repository of health and genetic information from half a million participants, to check whether polygenic risk scores provided added value. They found that the higher the polygenic risk score, the more likely it was that a person had, indeed, developed coronary heart disease or had a stroke.
In another part of that study, the researchers estimated how much better the polygenic risk scores would be at guiding treatment than standard risk assessments. First, they created a hypothetical population of 100,000 people ages 40 to 75, with the age, sex and cardiovascular disease characteristics of the general United Kingdom population. Then they imagined that all the people who got either a high or medium cardiovascular disease risk score based on standard factors also had a polygenic risk score calculated. Finally, of those people, they assumed that everyone whose genetics indicated high risk would be treated with cholesterol-busting statins.
The result: The team’s number crunching showed that more people would be identified as high risk and receive statins, and this could prevent 7 percent more heart attacks than standard screening alone. That could lead to a significant saving of lives.
Wanted: More non-Europeans
Despite this potential, polygenic risk scores today have a huge problem: They work best for people of European ancestry, as Martin and colleagues described in the Annual Review of Biomedical Data Science. Europeans make up about 16 percent of the world population but nearly 80 percent of participants in genome-wide association studies. That means the genetic variants Europeans tend to possess have an outsize influence on polygenic risk score algorithms.
But the genetic tags that point to disease in people of European descent might not work the same way in people of different ancestries. This goes back to the evolution of humankind: While Homo sapiens arose in Africa, over time different populations evolved different sets of small variants particular to each group. The genetic tags that exist in people of European descent might not exist in other groups — or they might be present, but without the same disease links.
For example, in a 2021 study of a polygenic risk score for heart-related traits such as diabetes or blood pressure, researchers found the scores were five times more accurate for European Americans compared to African Americans, and 20 times better for European Americans than for sub-Saharan Africans.
The fact that people of non-European descent are likely to get less accurate results from a polygenic risk score means that if score designers aren’t careful, their results could exacerbate existing health disparities, says Ryan Bogdan, a psychology researcher at Washington University in St. Louis.
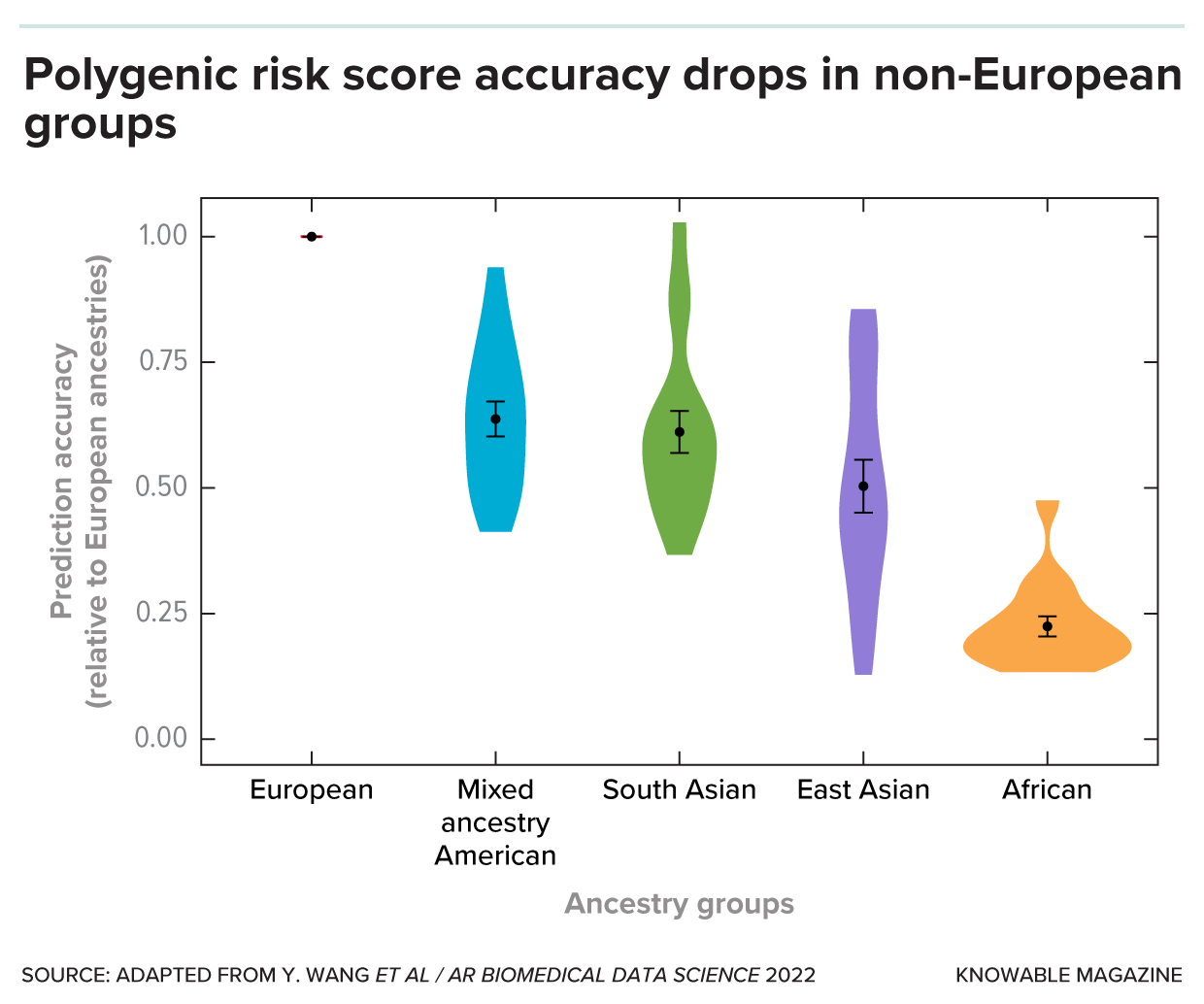
This graph shows how well polygenic risk scores work for different demographic groups, all compared to the scores’ performance among Europeans. Thus, the performance for Europeans is 1. The shapes are violin plots, which represent the distribution of the performance of scores for each ethnic group. Polygenic risk scores for all of the non-European groups are less accurate than they are for Europeans.
The solution is straightforward in theory, if difficult to put into practice: Recruit more diverse people for the genome-wide association studies that underlie polygenic risk scores. “The big push right now is trying to get more ancestrally diverse cohorts,” says Bogdan.
Reporting in 2023, Patel and colleagues did just that, incorporating genome-wide data from five different ancestries into a new polygenic risk score for coronary artery disease. The new one, they found, worked better for people of Hispanic, Asian and African ancestry. The new score also worked better for Europeans. That’s because there may be genetic variants linked to risk that are rare in Europeans or other ethnicities, but more likely to be found in people of more recent African ancestry. The more complete the data feeding into polygenic risk scores, the more accurate the scores that come out — for everyone.
Several initiatives are working to improve genetic diversity. Africa itself is the best place to look, because the people there contain the most genetic diversity. “When you research Africa, it’s the whole of humankind,” says Ananyo Choudhury, a bioinformaticist at the University of the Witwatersrand in Johannesburg.
Yet the challenges of building the infrastructure for widespread genetic studies in Africa have made it difficult to fulfill the continent’s genetic promise. H3Africa, a 10-year, $176 million project seeking genetic variants linked to diseases, wrapped up in 2022 with genetic data from 50,000 research participants. That’s a paltry number compared to genome-wide association studies performed elsewhere, Choudhury says.
But the work is continuing. For example, Meharry Medical College in Nashville, Tennessee, recently announced a five-year project to collect DNA from at least 500,000 people of African Ancestry, in both the United States and on the African continent. And the US-based All of US Research Program aims to collect genetic information from at least 1 million diverse individuals. Over the project’s first five years, it has accumulated genetic data from more than 312,000 people. That should be enough to improve risk scores for common conditions in people of various ethnicities, says Julia Moore Vogel, senior program director of the project’s Participant Center at Scripps Research in La Jolla, California.
With additional investment in research, polygenic risk scores may someday fulfill the promise of the human genome: personalized recommendations for even common, multifactorial conditions, for people of any ancestry, and thus the opportunity for everyone to be as healthy as their individual DNA will allow.
All the hurdles haven’t dampened scientist’s enthusiasm for the enormous potential of polygenic risk analysis. Watch this space: The field, says Martin, “is evolving extremely rapidly.”
10.1146/knowable-011724-2
TAKE A DEEPER DIVE | Explore Related Scholarly Articles